2.4 Adjacency matrix
The adjacency matrix of a network that has nodes has rows and columns. If there is a link from node to node , then . If node and node are not connected, then .
For an undirected network, the link has two representations: = . That’s why the adjacency matrix of an undirected network is always symmetric.
Lets have a look at the adjacency matrix of g1 and g2.
For g1:
par(mar = c(1, 1, 1, 1))
plot(g1)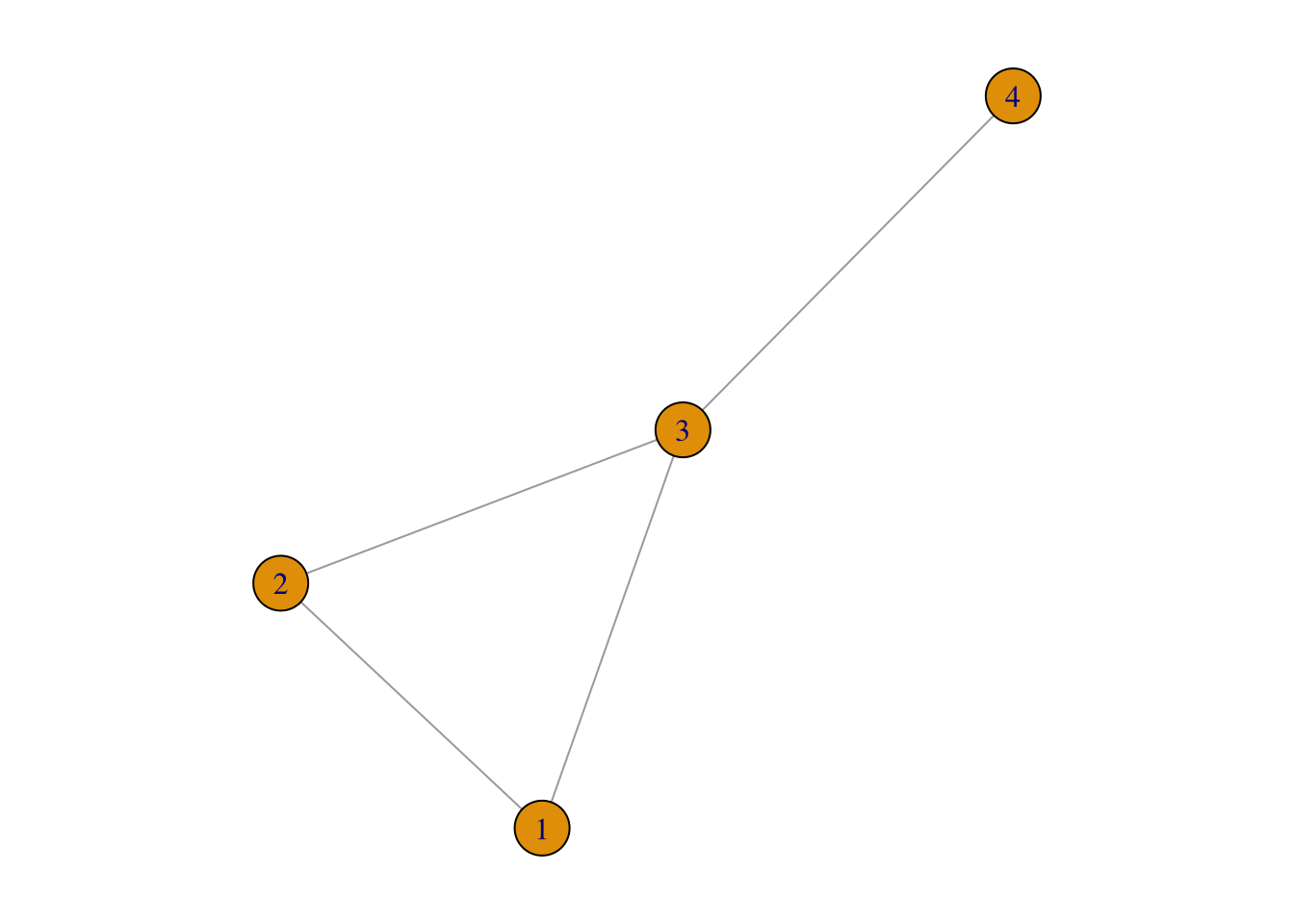
as_adjacency_matrix(g1)## 4 x 4 sparse Matrix of class "dgCMatrix"
##
## [1,] . 1 1 .
## [2,] 1 . 1 .
## [3,] 1 1 . 1
## [4,] . . 1 .For g2:
par(mar = c(1, 1, 1, 1))
plot(g2)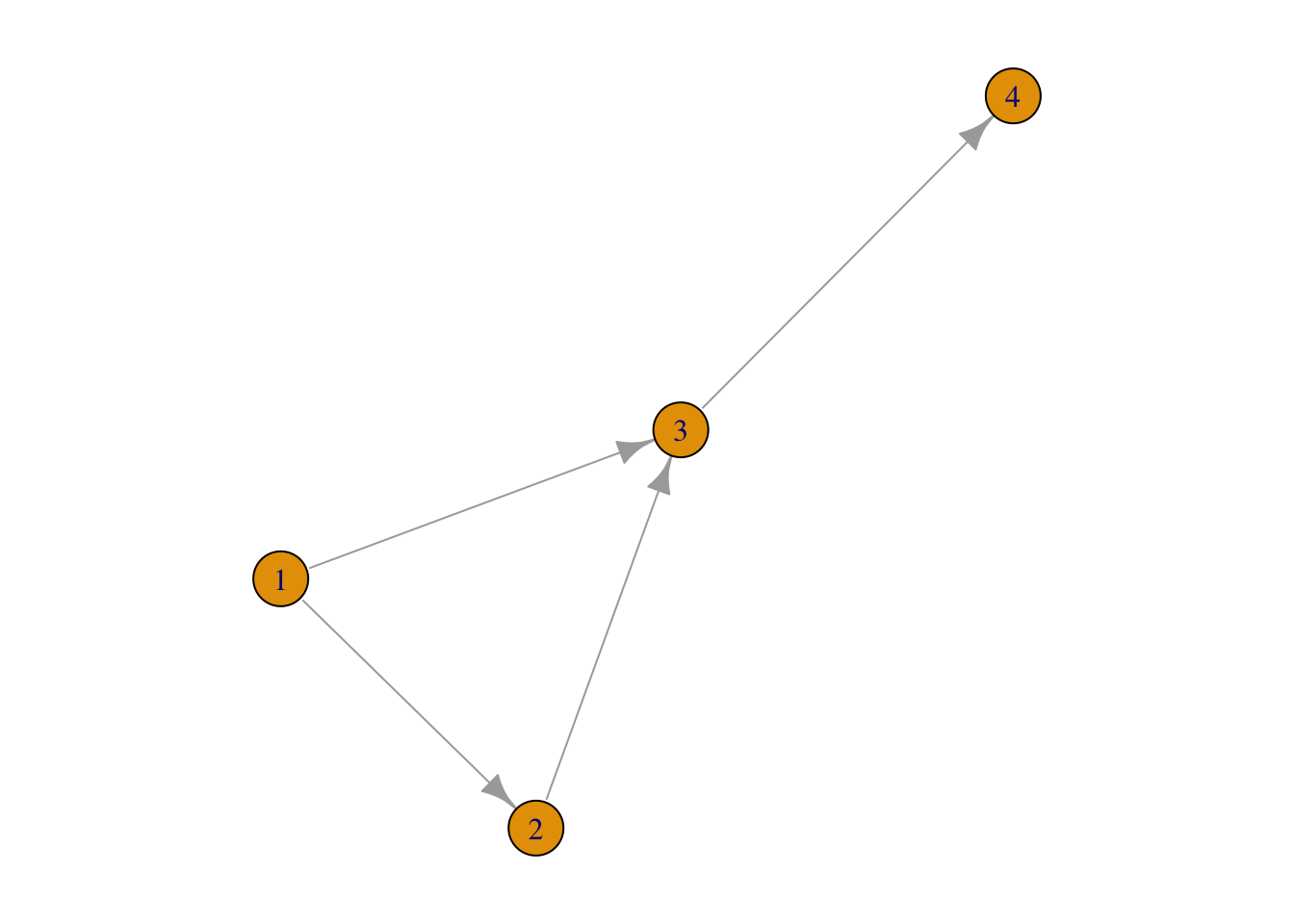
as_adjacency_matrix(g2)## 4 x 4 sparse Matrix of class "dgCMatrix"
##
## [1,] . 1 1 .
## [2,] . . 1 .
## [3,] . . . 1
## [4,] . . . .Each dot in the matrix means . The adjacency matrices here did not label the column name. To better understand it, you can imagine that the columns are also labeled as [1, ], [2, ], [3, ] and [4, ] as the rows are.